Entering edit mode
Hi, I'm trying to extract a significant gene list from a specific GO number. This is my code and all I get is an empty list. This empty list is under the title "Values" and it is a "character (empty).
goID <- "GO:0003677"
gene.universe <- genes(GOdataUp.res2.2)
go.genes <- genesInTerm(GOdataUp.res2.2, goID)[[2]]
sig.genes <- sigGenes(GOdataUp.res2.2)


Hi James, First thanks for the quick response! My problem is to produce a table of the significant LOC that belong to a significant GO number that has had a change in expression. I am trying to find this because there is a problem that is the same table as you entered above (In my code the table is called: "allResUp.res2.2"), written that the same GO number has 10 significant genes and in practice in the LOC table (In my code the table is called: "Final.loc.go.up.res2") there are more than 10 genes (there are actually 23 LOC) under the same GO. I wanted to ask if you have an idea for a command (in R software) in which I would be able to produce only the significant genes under the same GO? I enclose my last two chunks in the code (only the Upp Regulated) where the tables I mentioned appear are created:
Step 11: my tray to combain geneID2GO2.table withe downgenes.res2.GO to get 1 file with LOC and GO
Step 12: New Enrichment
To illustrate the problem, I also enclose a picture of the two tables facing each other: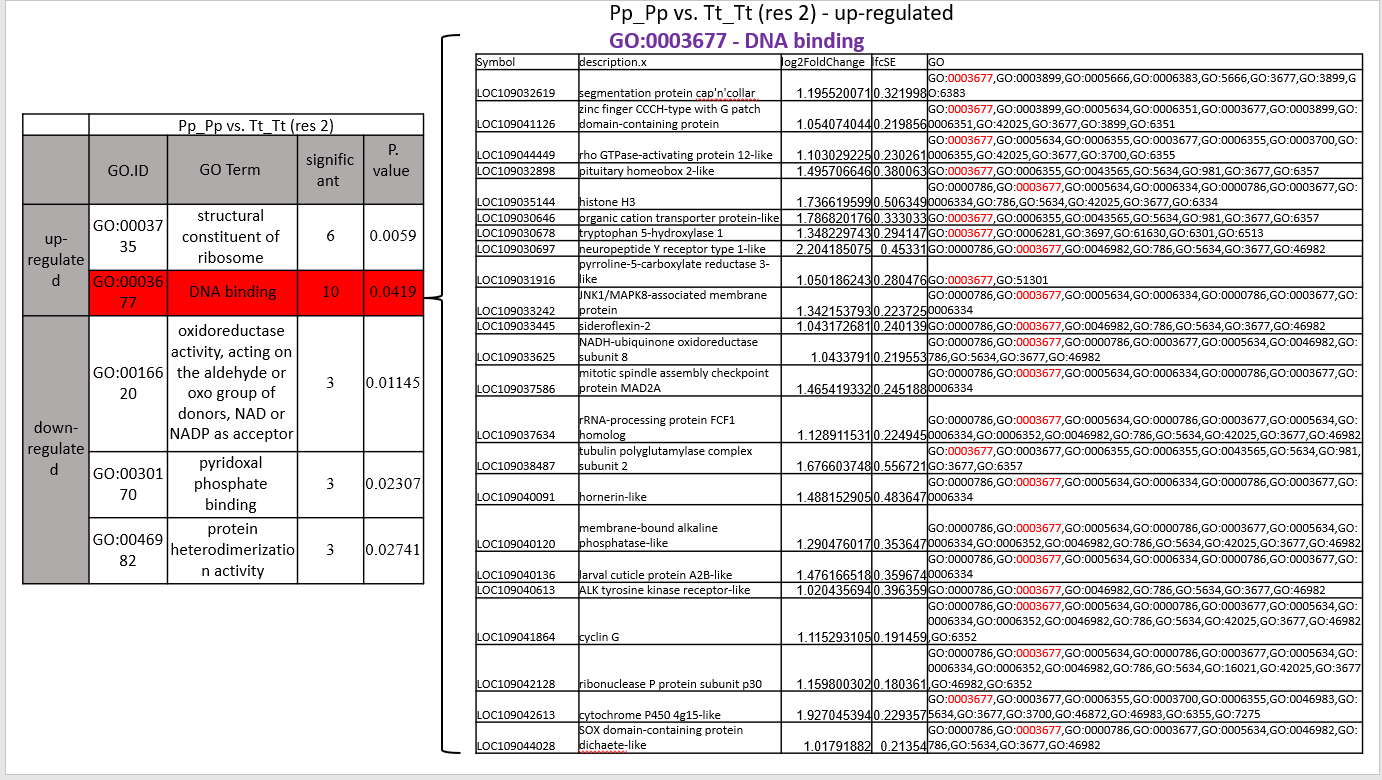
I do have a command to do that. It's the last line of the answer I already gave.
Hi James, I try your command and now my problem is that after I prepper the "sel.terms" I gett difrent GO namber then the GO namber I gett from the "allResUp.res2.2" table. Even the specific GO nember (GO:0003677) that I'm looking for is not apper in this list. The "GOdataUp.res2.2" is a larg topGOdate (2.8 MB). What I`m doing wrong?
Just to be precise, the "allResUp.res2.2" table is attached :
Many thanks in advance!!
Hi James, Update from now on is that I succeeded to create the correct table, but now my problem is that the last two lines in your code do not work. I got "character(0)" I need to find only the 10 significant genes that belong to this GO number (GO:0003677), maybe you have an idea of how to solve the problem? Attached is the latest code:
I think you are confusing the example data and your own. The goal of example data is to provide you with workable examples that you can extrapolate to your own data. It's not intended to be used directly with your data. You need to use the example to figure out how to make the same thing as the
sgvector using your own data.