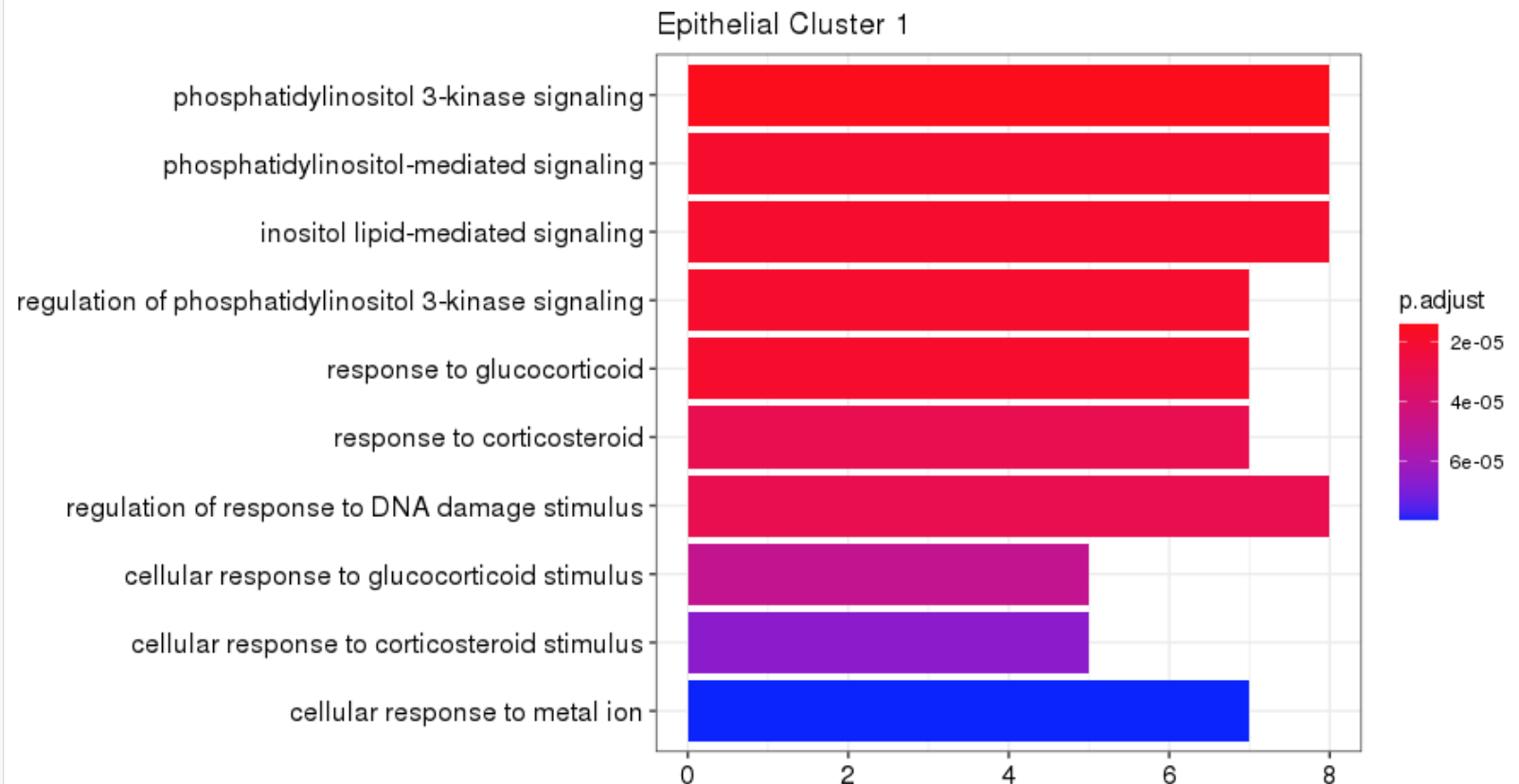
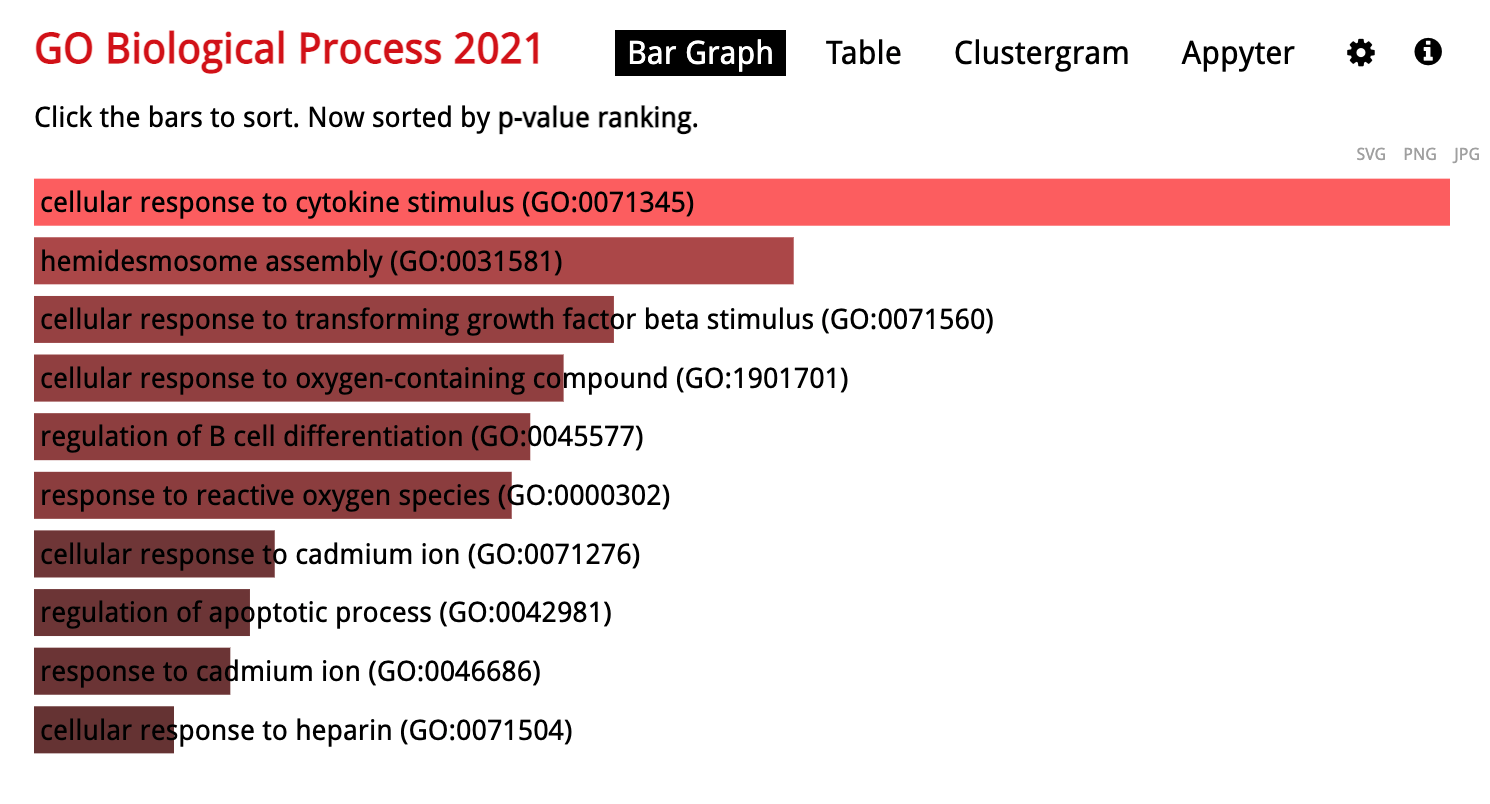
Hi there,
I have am getting different outputs after running enrichGO on cluster profiler when I put the same genes into enrichR (by Maayan Lab) website. Example here using Biological Process 2021
I have also tried other GO terms (Cellular Component and Molecular Funtion giving the same discordant results)
Code should be placed in three backticks as shown below
input_marker_files <- 'input_file'
for(i in 1:1){
markerFile <- paste0(input_marker_files, i,'.tsv')
marker.gene.clust[[i]] <- read.table(markerFile)
marker.gene.clust.filt <- marker.gene.clust[[i]][marker.gene.clust[[i]]$summary.logFC>1.5,]
#print(markerFile)
gene <- marker.gene.clust.filt$Symbol
gene.df <- bitr(gene, fromType = "SYMBOL",
toType = c("ENSEMBL", "ENTREZID"),
OrgDb = org.Hs.eg.db)
ego <- enrichGO(gene = gene.df$ENSEMBL,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05)
p2 <- barplot(ego, showCategory=10) +
theme(axis.text.y= element_text(size=12)) +
scale_y_discrete(guide = guide_axis(n.dodge=1)) +
ggtitle(paste0("Epithelial Cluster ", i))
p2
Thanks