Hi,
I am analyzing a NanoString dataset where the metadata look as below:
The factor of interest is the "group" and both groups are found in each mouse ("mouseID") (paired samples). I used DESeq2 for the analysis and the PCA on the normalized data looks like this:
It seems like there is a batch coming from the animals in PC1, although PC2 mildly separates the groups. I have set my design as:
~mouseID+group to account for differences between animals in my downstream DGE analysis. I wanted to ask whether you think
I should perform a stronger batch correction (e.g. limma). Also, I read a recent article on NanoString analysis where RUVg() was
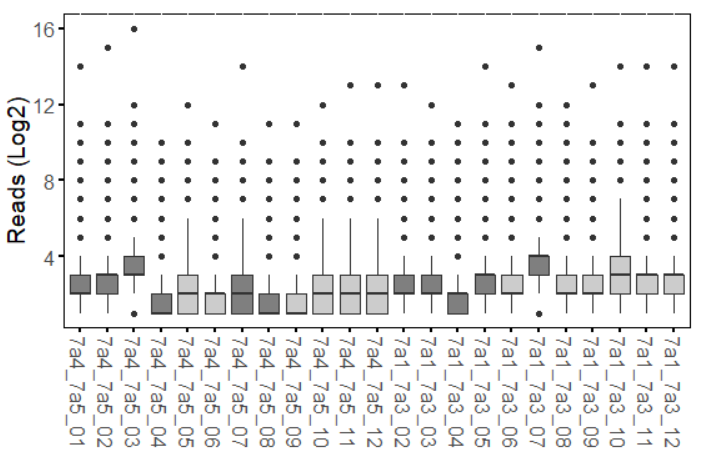
used for normalization. Below I attach how the samples look before and upon normalization with DESeq2 (counts()).
Do you reckon I should use RUVseq offset to normalize the samples?
Thanks in advance!

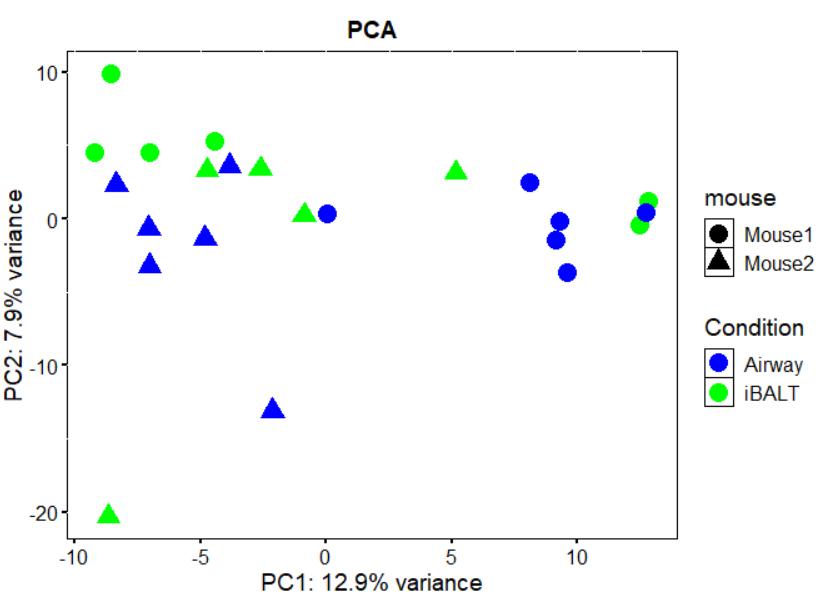
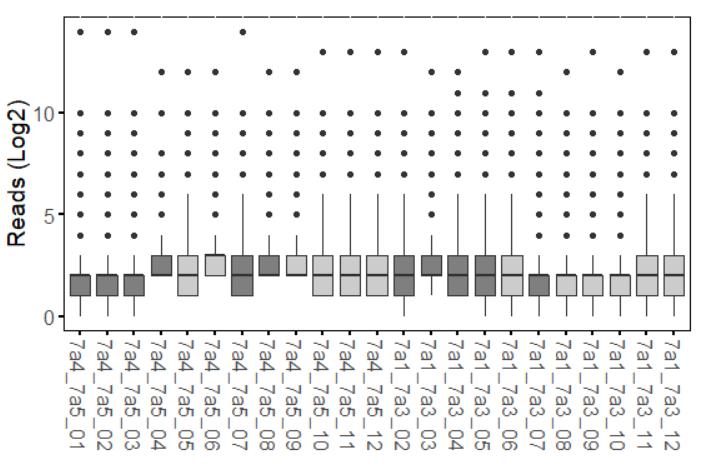

PC1 is only 12% of the variance? That's not a lot. It looks to me like none of your experimental conditions had much effect at all.