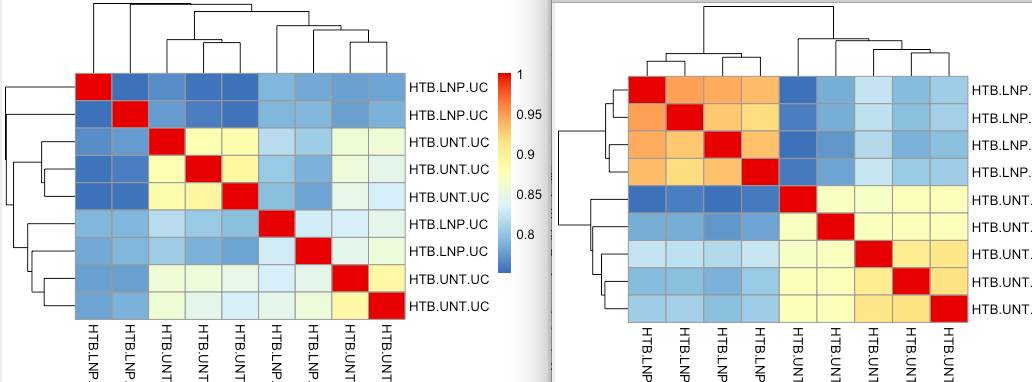
I was trying to plot a sample heatmap and we usually do it with VST count data, but I was playing with the TPM data today and found that the heatmap generated by VST TPM data has a much better clustering of untreated and treated samples.
I also tried plotting a PCA plot with VST TPM and I also get a very good grouping. I checked the standard deviation against the means of all genes and found that the VST TPM data became more homoscedastic than the VST count data.
I wonder if It is okay to use VST TPM data for heatmap and PCA, and what is the possible explanation why I got a better grouping result from VST TPM data.
Note that since the DESeqDataSet object expects integers, I rounded the TPM to the nearest integers before doing the transformaion.


Should I expect the two PCA plots to be the same when using the same ntop argument? I tried different ntop but did not get the same result. I also wonder about the sample heatmap, can VST TPM data be used for this or is it obviously wrong? or does it make sense to even use VST on TPM data? What happens to the TPM when I use VST that is different from using it on count data. Below are the PCA plots and sample heatmaps of VST count (left) and VST TPM (right). The sample heatmap of VST TPM has a better grouping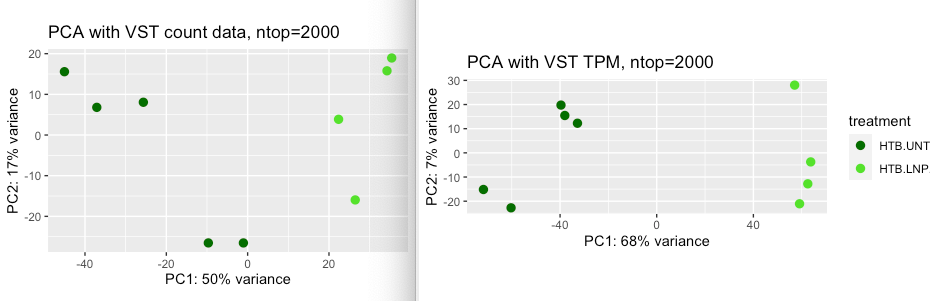
"Should I expect the two PCA plots to be the same when using the same ntop argument?"
No definitely not identical, because you are providing different input data.
But the two PCA plots look qualitatively similar to me. PC1 separates the samples by treatment and PC2 separates two UNT samples from three other UNT samples, maybe a mini-batch effect there.
I think what's happening is that genes which are DE across treatment have higher variance in the VST TPM dataset for whatever reason, and so are included more often in the top 2000 genes.
Can you try another thing: what about rlog on count data, using
rlog(dds, blind=FALSE)?Hello again, Thank you very much for your reply. I had used rlog on the count data and got a good grouping on the sample heat map and an almost identical PCA plot to the VST count data.
I would appreciate it if you could guide me about the heatmap. I initially planned to use VST TPM for heatmap since it gives better sample clustering (separate UNT untreated and LNP treated). Can this be done is it obviously wrong? Does it make sense to even use VST on TPM data? What happens to the TPM when I use VST that is different from using it on count data since the heatmap groups better with VST TPM.
I included the SD&mean plots below which I used to check the homoskedasticity of the VST count, VST TPM, and rlog count data. The plots here includes samples from one cell line. The VST TPM looks more homoskedastic when I included all samples from all cell lines.
We were struggling with justifying using the VST TPM for a sample heatmap and would like to make sure to use the data correctly for a manuscript.
The DESeq2 VST in
vst()doesn't make sense on TPM because it is designed for NB distributed count data. The TPM are not close to NB (negative binomial).To me, plots 1 and 3 (VST on counts and rlog) look good (don't worry about the dip down to 0 on the far left side, this is unavoidable as the counts -> 0 so must the SD of the VST data).
The VST on TPM has a strong trend of SD with mean TPM (first up then down, peaking at ~10^0.5), and I would not recommend to use this approach.