macbook m1 R studio install deseq2 dependent package genefilter failed
r studio
BiocManager::install('genefilter')
#result show
'getOption("repos")' replaces Bioconductor standard repositories, see '?repositories' for details
replacement repositories:
CRAN: https://mirrors.tuna.tsinghua.edu.cn/CRAN/
Bioconductor version 3.13 (BiocManager 1.30.16), R 4.1.0 (2021-05-18)
Installing package(s) 'genefilter'
Package which is only available in source form, and may need compilation of
C/C++/Fortran: ‘genefilter’
Do you want to attempt to install these from sources? (Yes/no/cancel) Yes
installing the source package ‘genefilter’
trying URL 'https://bioconductor.org/packages/3.13/bioc/src/contrib/genefilter_1.74.0.tar.gz'
Content type 'application/x-gzip' length 805682 bytes (786 KB)
==================================================
downloaded 786 KB
* installing *source* package ‘genefilter’ ...
** using staged installation
** libs
clang++ -arch arm64 -std=gnu++14 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c half_range_mode.cpp -o half_range_mode.o
clang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c init.c -o init.o
clang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c nd.c -o nd.o
clang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c pAUC.c -o pAUC.o
clang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c rowPAUCs.c -o rowPAUCs.o
clang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c rowttests.c -o rowttests.o
/opt/R/arm64/bin/gfortran -mtune=native -fno-optimize-sibling-calls -fPIC -Wall -g -O2 -c ttest.f -o ttest.o
make: /opt/R/arm64/bin/gfortran: No such file or directory
make: *** [ttest.o] Error 1
ERROR: compilation failed for package ‘genefilter’
* removing ‘/Library/Frameworks/R.framework/Versions/4.1-arm64/Resources/library/genefilter’
The downloaded source packages are in
‘/private/var/folders/bt/t904ydv902d4lqg2_jxf_n600000gn/T/Rtmpz5VVBy/downloaded_packages’
Warning messages:
1: In .inet_warning(msg) :
unable to access index for repository https://bioconductor.org/packages/3.13/bioc/bin/macosx/big-sur-arm64/contrib/4.1:
cannot open URL 'https://bioconductor.org/packages/3.13/bioc/bin/macosx/big-sur-arm64/contrib/4.1/PACKAGES'
2: In .inet_warning(msg) :
unable to access index for repository https://bioconductor.org/packages/3.13/data/annotation/bin/macosx/big-sur-arm64/contrib/4.1:
cannot open URL 'https://bioconductor.org/packages/3.13/data/annotation/bin/macosx/big-sur-arm64/contrib/4.1/PACKAGES'
3: In .inet_warning(msg) :
unable to access index for repository https://bioconductor.org/packages/3.13/data/experiment/bin/macosx/big-sur-arm64/contrib/4.1:
cannot open URL 'https://bioconductor.org/packages/3.13/data/experiment/bin/macosx/big-sur-arm64/contrib/4.1/PACKAGES'
4: In .inet_warning(msg) :
unable to access index for repository https://bioconductor.org/packages/3.13/workflows/bin/macosx/big-sur-arm64/contrib/4.1:
cannot open URL 'https://bioconductor.org/packages/3.13/workflows/bin/macosx/big-sur-arm64/contrib/4.1/PACKAGES'
5: In .inet_warning(msg) :
unable to access index for repository https://bioconductor.org/packages/3.13/books/bin/macosx/big-sur-arm64/contrib/4.1:
cannot open URL 'https://bioconductor.org/packages/3.13/books/bin/macosx/big-sur-arm64/contrib/4.1/PACKAGES'
6: In .inet_warning(msg) :
installation of package ‘genefilter’ had non-zero exit status
the error showed: make: /opt/R/arm64/bin/gfortran: No such file or directory. so I downloaded gfortran in terminal
macbook terminal
neurox@NeuroX ~ % cat ./.bash_profile
export gfortran=/usr/local/gfortran/bin
export PATH=$PATH:$gfortran
neurox@NeuroX ~ % source ./.bash_profile
neurox@NeuroX ~ % gfortran -v
使用内建 specs。
COLLECT_GCC=gfortran
COLLECT_LTO_WRAPPER=/usr/local/gfortran/libexec/gcc/x86_64-apple-darwin15/6.1.0/lto-wrapper
目标:x86_64-apple-darwin15
配置为:../gcc-6.1.0/configure --prefix=/usr/local/gfortran --with-gmp=/Users/fx/devel/gcc/deps-static/x86_64 --enable-languages=c,c++,fortran,objc,obj-c++ --build=x86_64-apple-darwin15
线程模型:posix
gcc 版本 6.1.0 (GCC)
Also, I add it to the r studio path
Sys.setenv(gfortran="/usr/local/gfortran/bin") then still failed
still get
make: /opt/R/arm64/bin/gfortran: No such file or directory
so i do this
# make link to /opt/R/arm64/bin
sudo ln -s /usr/local//gfortran/bin/gfortran /opt/R/arm64/bin
finally
BiocManager::install('genefilter')
make: /opt/R/arm64/bin/gfortran: Permission denied
upset!!! i need help thanks!


Hi soda, I've met the same problem with you. I fail to install the "genefilter" package with my M1 MacBook arm64 Studio I've tried several ways I could find to solve this problem, including root or modify the "/.config/Rstudio" file, and it still showed
make: /opt/R/arm64/bin/gfortran: Permission deniedSince you had solved this problem by yourself (download gfortran in path /opt/R/arm64/bin), I did as the instructions. However, I could not download it in the correct path (it would be downloaded in "/User/id" instead of "/opt/R/arm64/bin") and could not open it with sudo tar fvxz even in the su root I really hope you can help me with this.Best regards Zhusheng Zhang
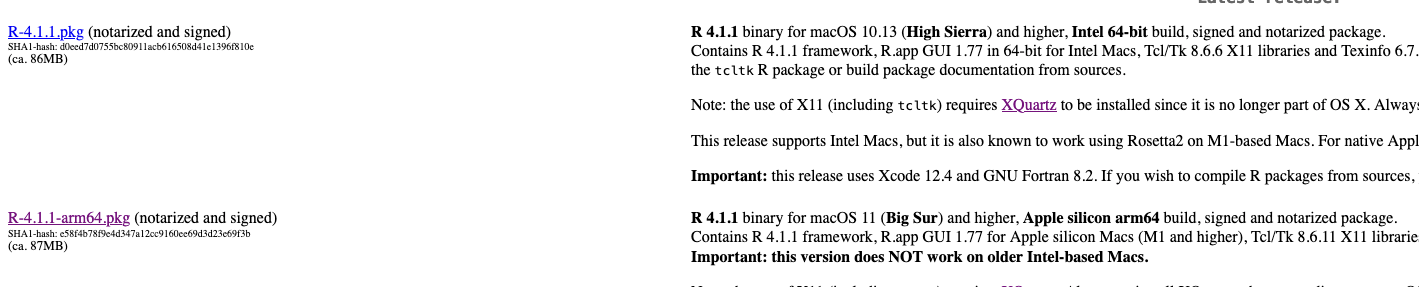
Hello Zhusheng Zhang, Except this problem, R arm64 has many other bugs.So I unload R arm64 ,then download 'Intel 64-bit build' version. Now, I have never encountered any problems about installing package. The following answer is the same as mine,you can read it.