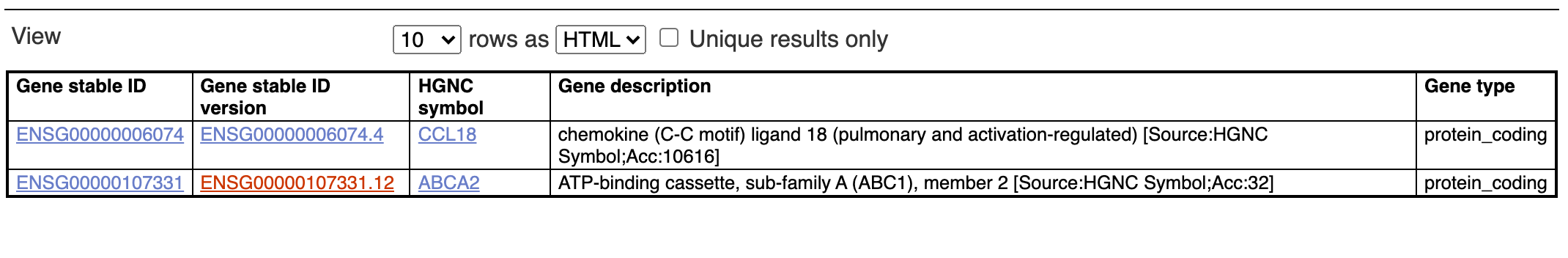
Here is a fun little problem. Here is an example gene: ENSG00000006074 - CCL18
I get the result expected on the ensembl website (including ENSG00000107331 as a positive control):
Here is the XML query from the biomart site (http://grch37.ensembl.org/index.html)
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE Query>
<Query virtualSchemaName = "default" formatter = "TSV" header = "0" uniqueRows = "0" count = "" datasetConfigVersion = "0.6" >
<Dataset name = "hsapiens_gene_ensembl" interface = "default" >
<Filter name = "ensembl_gene_id" value = "ENSG00000006074,ENSG00000107331"/>
<Attribute name = "ensembl_gene_id" />
<Attribute name = "ensembl_gene_id_version" />
<Attribute name = "hgnc_symbol" />
<Attribute name = "description" />
<Attribute name = "gene_biotype" />
</Dataset>
</Query>
However, let's try in R (using either biomaRt or do it ourself):
fullXmlQuery <- "<?xml version='1.0' encoding='UTF-8'?><!DOCTYPE Query><Query virtualSchemaName = 'default' uniqueRows = '0' count='' datasetConfigVersion='0.6' header='1' formatter='TSV' requestid='biomaRt'> <Dataset name = 'hsapiens_gene_ensembl' interface = 'default'><Attribute name = 'ensembl_gene_id'/><Attribute name = 'description'/><Attribute name = 'hgnc_symbol'/><Attribute name = 'gene_biotype'/><Filter name = 'ensembl_gene_id' value = 'ENSG00000006074,ENSG00000107331' /></Dataset></Query>"
res <- httr::POST(url = "https://apr2019.archive.ensembl.org:443/biomart/martservice",
body = list('query' = fullXmlQuery),
config = httr::config())
httr::content(res)
This will only return results for ENSG00000107331 and never ENSG00000006074.
[1] "Gene stable ID\tGene description\tHGNC symbol\tGene type\nENSG00000107331\tATP binding cassette subfamily A member 2 [Source:HGNC Symbol;Acc:HGNC:32]\tABCA2\tprotein_coding\n"
I also see this issue if you try a service like g:Profiler (https://biit.cs.ut.ee/gprofiler/convert):
Here is the biomaRt package query too:
ens_version <- biomaRt::useEnsembl(biomart = 'genes',
dataset = 'hsapiens_gene_ensembl',
version = 96)
# return
biomaRt::getBM(attributes = c("ensembl_gene_id",
"description",
"hgnc_symbol",
"gene_biotype"), #,"entrezgene"
filters = 'ensembl_gene_id',
values = "ENSG00000006074",
mart = ens_version,
uniqueRows = FALSE)
Returns:
[1] ensembl_gene_id description hgnc_symbol gene_biotype
<0 rows> (or 0-length row.names)
Is this an encoding issue or a known issue due to versions? Note that I have tried several ensembl versions. I have not exhaustively searched for all genes that have an issue.

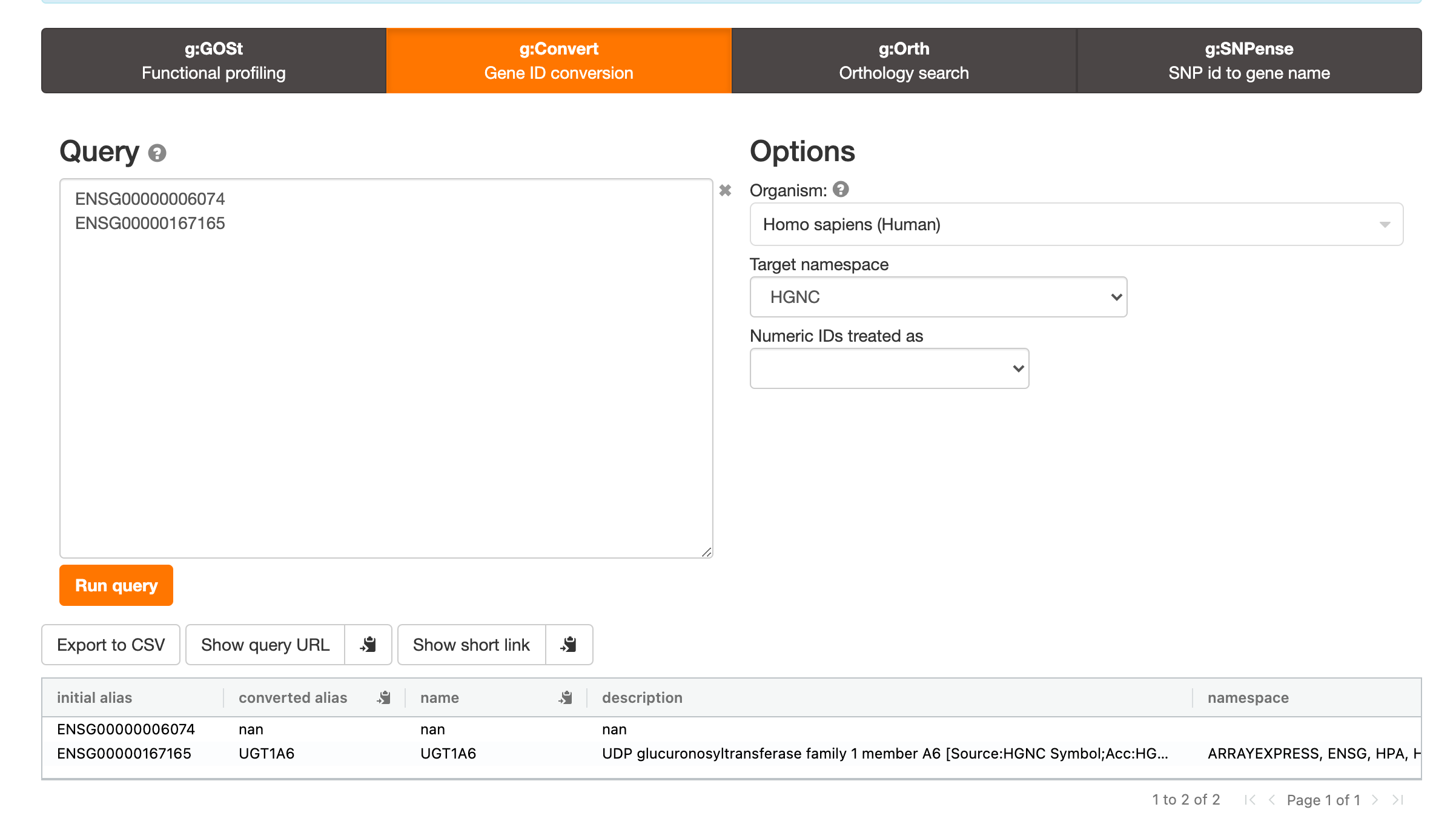
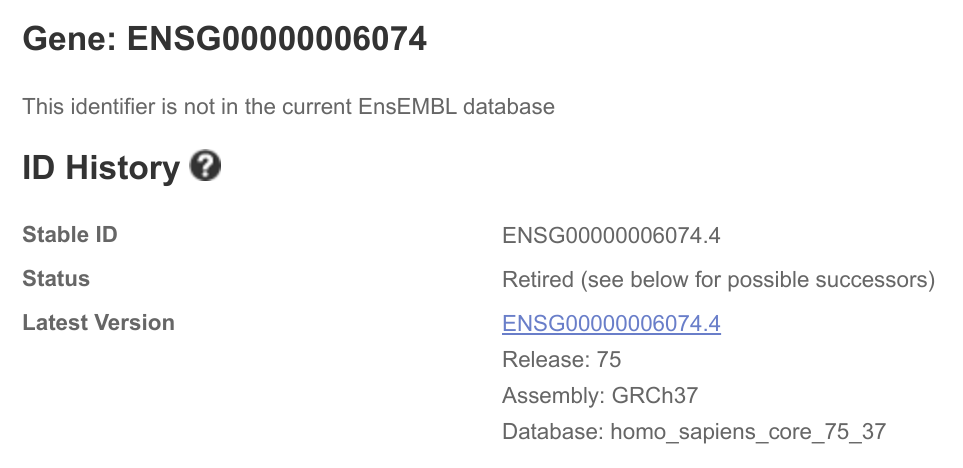

It appears that this is an GRCh37 versus GRCh38 issue. I want to map to hg19/GRCh37 IDs but the default for these services are GRCh38.
If anyone else finds this, you can change assembly in biomaRt like this:
The url for queries is: "https://grch37.ensembl.org:443/biomart/martservice"