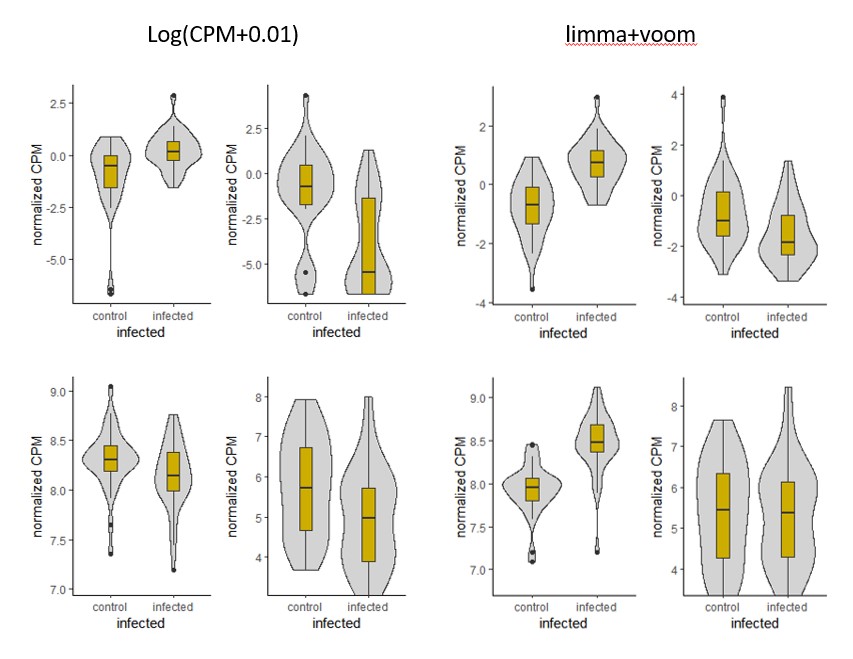
Hello, I'm encountering an odd situation where the effect of stimulation on a gene's expression seems to flip sign after applying limma+voom. We noticed this when the data plotted as log2(cpm+0.01) seemed to have an opposite effect size as the results of lmFit. The gene is pretty highly expressed (log2(CPM) ~ 8) with 140-4000 reads per sample and time point. For comparison, I'm showing below the log2(cpm+0.01) on the left for 4 genes and the voom normalized data in v$E for the same 4 genes on the right; it's the gene in the bottom left of both plots that shows a reversal in sign (I know v$E isn't a variable that's supposed to be plotted, but it illustrates the difference between CPM and voom-normalized data which is also reflected in the effect size from lmFit). Further below, I've also included my code for this analysis. We're modeling an effect of treatment, while controlling for individual (each individual is sampled twice, once in infected and once in control).
Are there any explanations for how voom would change the sign of an effect like this compared to the cpm data? Thank you very much for your help!
# calculate CPM
cpm <- counts; for (i in 1:66) {cpm[,i] <- (counts[,i]*1e6)/metadata$reads[i]}; rm(i)
# normalize with voom
y <- DGEList(counts = counts, lib.size = metadata$reads, samples = metadata[,2:3], genes = row.names(counts))
y <- calcNormFactors(y)
design <- model.matrix(~ 1 + stimulated + indiv_names)
colnames(design)[1:2] <- c("intercept", "stimulated")
v = voom(y, design, plot = T, save.plot = T)
# linear model using lmfit
fit = lmFit(v, design)
contr.matrix <- makeContrasts(
Stimulation = stimulated,
levels=colnames(design)
)
fit2 <- contrasts.fit(fit = fit, contrasts = contr.matrix)
fit2 = eBayes(fit2, robust=T)
res_lmfit <- topTreat(fit2, coef=1, n=Inf)
sessionInfo()
R version 4.0.2 (2020-06-22)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252 LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ggplot2_3.3.2 edgeR_3.30.3 limma_3.44.3 lme4_1.1-27.1 Matrix_1.3-4
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 compiler_4.0.2 pillar_1.4.6 nloptr_1.2.2.2 tools_4.0.2
[6] statmod_1.4.35 boot_1.3-25 digest_0.6.25 evaluate_0.14 lifecycle_0.2.0
[11] tibble_3.0.3 gtable_0.3.0 nlme_3.1-148 lattice_0.20-41 pkgconfig_2.0.3
[16] rlang_0.4.10 yaml_2.2.1 xfun_0.18 withr_2.3.0 dplyr_1.0.2
[21] knitr_1.30 generics_0.1.0 vctrs_0.3.6 tidyselect_1.1.0 locfit_1.5-9.4
[26] grid_4.0.2 glue_1.4.2 R6_2.4.1 rmarkdown_2.5 minqa_1.2.4
[31] farver_2.0.3 purrr_0.3.4 magrittr_1.5 scales_1.1.1 htmltools_0.5.1.1
[36] ellipsis_0.3.1 MASS_7.3-51.6 splines_4.0.2 colorspace_1.4-1 labeling_0.4.2
[41] munsell_0.5.0 crayon_1.4.1


Cross posted to Biostars https://www.biostars.org/p/9490039/