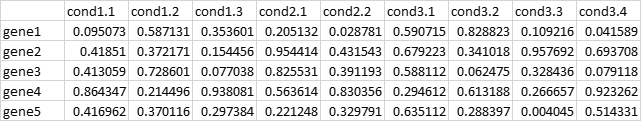
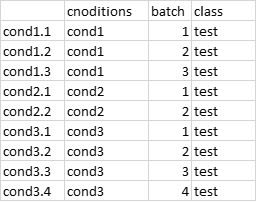
I have a data matrix of 3264 by 23. The rows are genes and columns are treatments. The treatments have an unequal number of replicates done in four batches. Examples of my files with random numbers are given below. My phenom file has 4 batches (6,6,6,5) and 11 conditions. The codes below are modified for the example data.
dt.matrix
phenom
batch1 <- phenom$batch
mod1 <- model.matrix(~conditions, data=phenom)
combt.p <- ComBat(dat=dt.matrix ,
mod=mod1 ,
batch=batch1 ,par.prior=TRUE, prior.plots=TRUE)
# Error output
Found4batches
Adjusting for 3 covariate(s) or covariate level(s)
Error in ComBat(dat = dt.matrix, mod = mod1, batch = batch1, par.prior = TRUE, :
At least one covariate is confounded with batch! Please remove confounded covariates and rerun ComBat
# Then I went into the ComBat codes and look for step-specific issues. I found that running codes below was causing issues.
(qr(design)$rank < ncol(design))
TRUE
#This output was supposed to give the result "FALSE". I even tried removing individual treatments and re-running the code but got the same result. Out of the 15 data matrices that I have, I got the same error for 4 matrices. I have no idea what is going wrong with my files or codes. Please help.
sessionInfo( )


Thank you James for your response. I tried
limmaand seems like my data matrix was valid for full rank.My stats knowledge is very limited, that is why I am struggling with this issue. Could you please guide me towards the right direction of what can be tested to get my data ready for comBat? Thank you.
The error you get from
ComBatcomes from the testAnd inside of
nonEstimableis, as one might expectWhere
xis your design matrix. That's the exact same test! So if you were getting an error fromComBatand now you aren't getting anything fromnonEstimable, the only possibility is that you are using different design matrices.Thank you, James. I was indeed using the wrong matrix. The
nonExtimabledid give me the column name which was causing the issue but I do not know how to fix it. Simply removing the column or the entire treatment set didn't fix the issue. I have reached to a stats professor here for help.