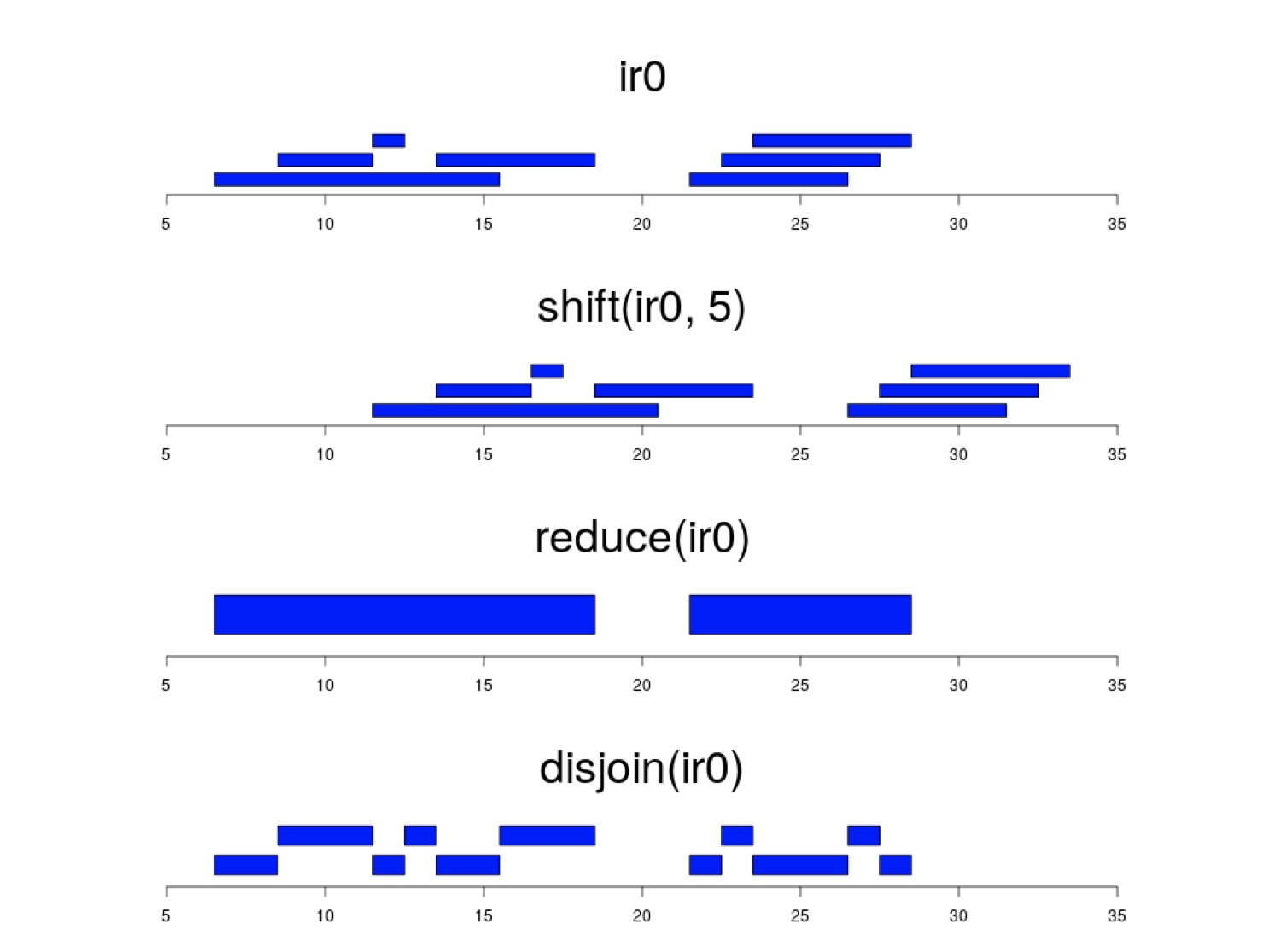
Every time I want to use ranges-related functions from the package GenomicRanges I need to explore docs again and again. Is this common to everyone? What's the best way to remember what GRanges functions (e.g. shift, reduce, join, disjoin, intersect, etc.) do? Like does anyone know if there's any pictorial representation or cheat sheet to remember GRanges functions? I have found one (attached here) in the package documentation. Any better than this?
`GenomicRanges` cheat sheet
1
Entering edit mode
2
Entering edit mode
Not pictoral, but I wrote up this table a while ago for quick reference:
https://github.com/mikelove/bioc-refcard/blob/master/README.Rmd#L149
I often now use and teach plyranges, as students are familiar with dplyr and the language adapts easily, including grouping and summarization.
https://sa-lee.github.io/plyranges/articles/an-introduction.html#arithmetic-on-ranges-1
shift(x, 5) is: shift_right(x, 5) or _upstream and _downstream can be used to move with respect to strand. The vignette above has more examples. Anchoring various points in the range and mutating the width is easy to write and read in my opinion.
Similar Posts
Loading Similar Posts
Traffic: 637 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thanks, Love. Good to know about plyranges. I will play with that.