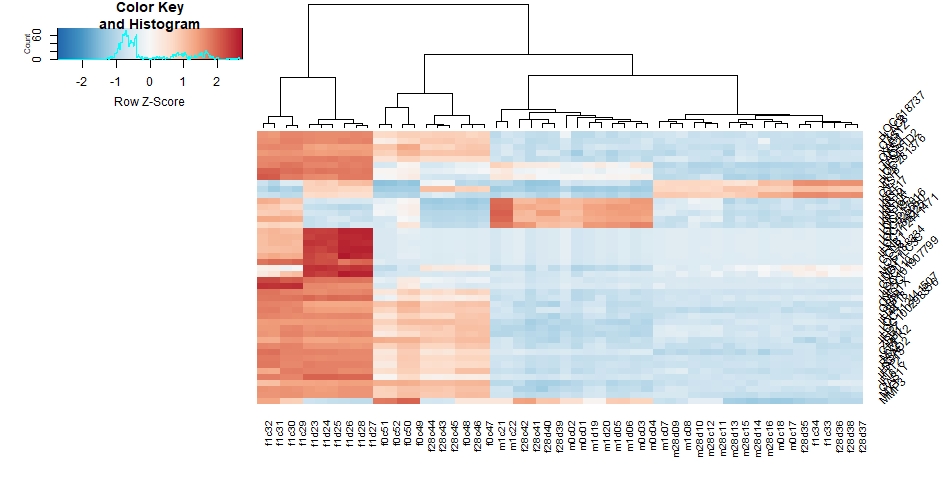
Entering edit mode
Hi,
I am trying to draw a heatmap for my 45 topvar gene by the use of heatmap.2, and when I set a srtRow=45 in my code(below):
heatmap.2( assay(rld)[ topVarGenes, ], srtRow=45, scale="row",trace="none", dendrogram="column",col = colorRampPalette( rev(brewer.pal(9, "RdBu")) )(255))
row names collided with each other in messy way.
would you please help me to solve this problem? (also I'm a newbie beginner using R Studio)
Thank you so much