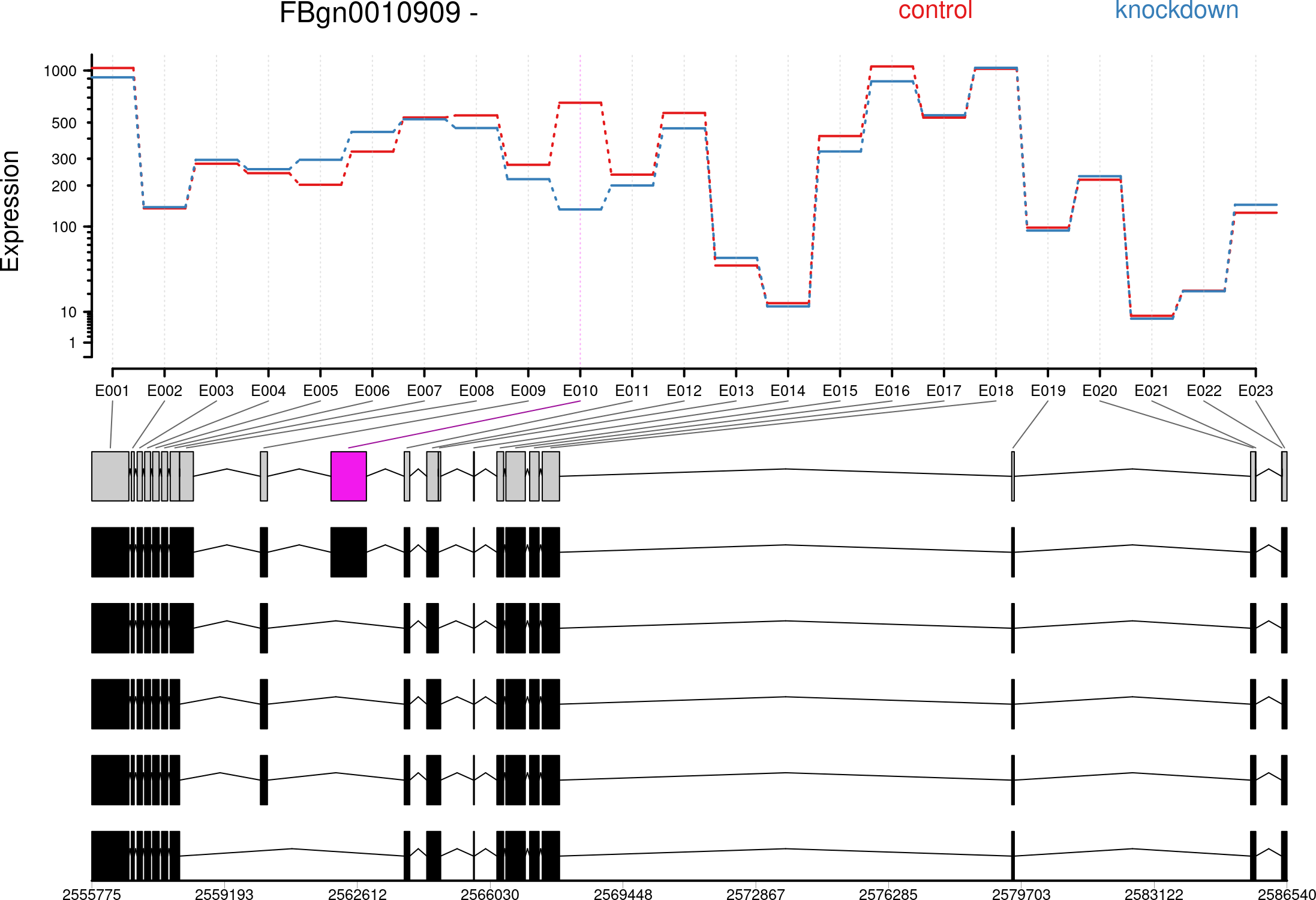
I have performed differential exon usage with DEXSeq using the corresponding vignette. It worked fine but now I want to investage a hit further, e.g. ENSG00000141510:E002 where E002 refers to an exon counting bin and not necessarily to an exon:
We need to “collapse” this information to define exon counting bins, i.e., a list of intervals, each corresponding to one exon or part of an exon. Counting bins for parts of exons arise when an exonic region appears with different boundaries in different transcripts.
Therefore, one can't simply map E002 to an Ensembl Exon ID and continue the this. I am wondering what is the easiest way to check whether in which transcripts E002 is included and in which region on the transcript or on the respective protein?


Thanks Alejandro, for the vignette, I think it will do what I want. I will give you feedback as soon as I have tried it.