Hello,
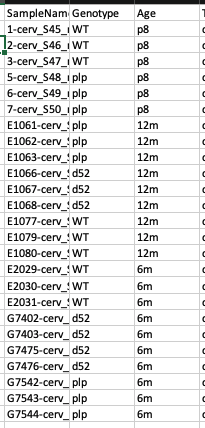
I am attempting to analyze expression differences between three genotypes of mice (1 wildtype, 2 mutated) at three different ages. I would like to compare expression at a particular age across the genotypes – for example, compare wildtype to d52 at age 6 months. I have attached my sample list for clarity:
However, because my model matrix is not full rank, I manually edited the matrix as per the vignette. I am still having trouble though because when I call resultsNames(dds), I am not getting the contrasts that I want.
ddsHTSeq <- DESeqDataSetFromHTSeqCount(sampleTable = sampletable,
directory = directory,
design = ~ 1)
ddsHTSeq_filtered <- ddsHTSeq[rowSums(counts(ddsHTSeq)) > 10, ]
custom.matrix <- model.matrix(~ Genotype + Age:Genotype, data = colData(ddsHTSeq_filtered))
all.zero <- apply(custom.matrix, 2, function(x) all(x==0))
idx <- which(all.zero)
custom.matrix <- custom.matrix[,-idx]
dds <- DESeq(ddsHTSeq_filtered, full = custom.matrix)
resultsNames(dds)
Output: "Intercept" "Genotypemdx" "GenotypeWT" "Genotyped52.Age6m" "Genotypemdx.Age6m" "GenotypeWT.Age6m" "Genotypemdx.Agep8" "GenotypeWT.Agep8"
I also attempted to extract contrasts using the following:
results(dds, contrast = c("Genotype", "WT", "mdx"))
But I receive the following error: only list- and numeric-type contrasts are supported for user-supplied model matrices
Any help would be greatly appreciated!!
sessionInfo( )
R version 3.6.2 (2019-12-12)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.4 purrr_0.3.4
[5] readr_1.4.0 tidyr_1.1.2 tibble_3.0.6 tidyverse_1.3.0
[9] ggplot2_3.3.3 pheatmap_1.0.12 apeglm_1.8.0 DESeq2_1.26.0
[13] SummarizedExperiment_1.16.1 DelayedArray_0.12.3 BiocParallel_1.20.1 matrixStats_0.58.0
[17] Biobase_2.46.0 GenomicRanges_1.38.0 GenomeInfoDb_1.22.1 IRanges_2.20.2
[21] S4Vectors_0.24.4 BiocGenerics_0.32.0
loaded via a namespace (and not attached):
[1] colorspace_2.0-0 ellipsis_0.3.1 htmlTable_2.1.0 XVector_0.26.0 base64enc_0.1-3
[6] fs_1.5.0 rstudioapi_0.13 farver_2.0.3 bit64_4.0.5 AnnotationDbi_1.48.0
[11] mvtnorm_1.1-1 lubridate_1.7.9.2 xml2_1.3.2 splines_3.6.2 cachem_1.0.4
[16] geneplotter_1.64.0 knitr_1.31 Formula_1.2-4 jsonlite_1.7.2 broom_0.7.4
[21] annotate_1.64.0 cluster_2.1.1 dbplyr_2.1.0 png_0.1-7 compiler_3.6.2
[26] httr_1.4.2 backports_1.2.1 assertthat_0.2.1 Matrix_1.3-2 fastmap_1.1.0
[31] cli_2.3.0 htmltools_0.5.1.1 tools_3.6.2 coda_0.19-4 gtable_0.3.0
[36] glue_1.4.2 GenomeInfoDbData_1.2.2 Rcpp_1.0.6 bbmle_1.0.23.1 cellranger_1.1.0
[41] vctrs_0.3.6 xfun_0.21 rvest_0.3.6 lifecycle_1.0.0 XML_3.99-0.3
[46] zlibbioc_1.32.0 MASS_7.3-53.1 scales_1.1.1 hms_1.0.0 RColorBrewer_1.1-2
[51] memoise_2.0.0 gridExtra_2.3 emdbook_1.3.12 bdsmatrix_1.3-4 rpart_4.1-15
[56] latticeExtra_0.6-29 stringi_1.5.3 RSQLite_2.2.3 genefilter_1.68.0 checkmate_2.0.0
[61] rlang_0.4.10 pkgconfig_2.0.3 bitops_1.0-6 lattice_0.20-41 labeling_0.4.2
[66] htmlwidgets_1.5.3 bit_4.0.4 tidyselect_1.1.0 plyr_1.8.6 magrittr_2.0.1
[71] R6_2.5.0 generics_0.1.0 Hmisc_4.4-2 DBI_1.1.1 pillar_1.4.7
[76] haven_2.3.1 foreign_0.8-75 withr_2.4.1 survival_3.2-7 RCurl_1.98-1.2
[81] nnet_7.3-15 modelr_0.1.8 crayon_1.4.1 jpeg_0.1-8.1 locfit_1.5-9.4
[86] grid_3.6.2 readxl_1.3.1 data.table_1.13.6 blob_1.2.1 reprex_1.0.0
[91] digest_0.6.27 xtable_1.8-4 numDeriv_2016.8-1.1 munsell_0.5.0


Thanks for the response! Would using the following
results(dds, contrast = list(c("GenotypeWT.Age6m","Genotypemdx.Age6m")))contrast the differences in the WT genotype and mdx genotype for the 6 month old cohort?I'm sorry I just don't have sufficient time to help with interpretation of results on the support site, and have to restrict myself to software related questions. I'd recommend to work with statistician to interpret and build the contrasts of interest.