I am trying to use pathview to do this (example from https://pathview.r-forge.r-project.org/):
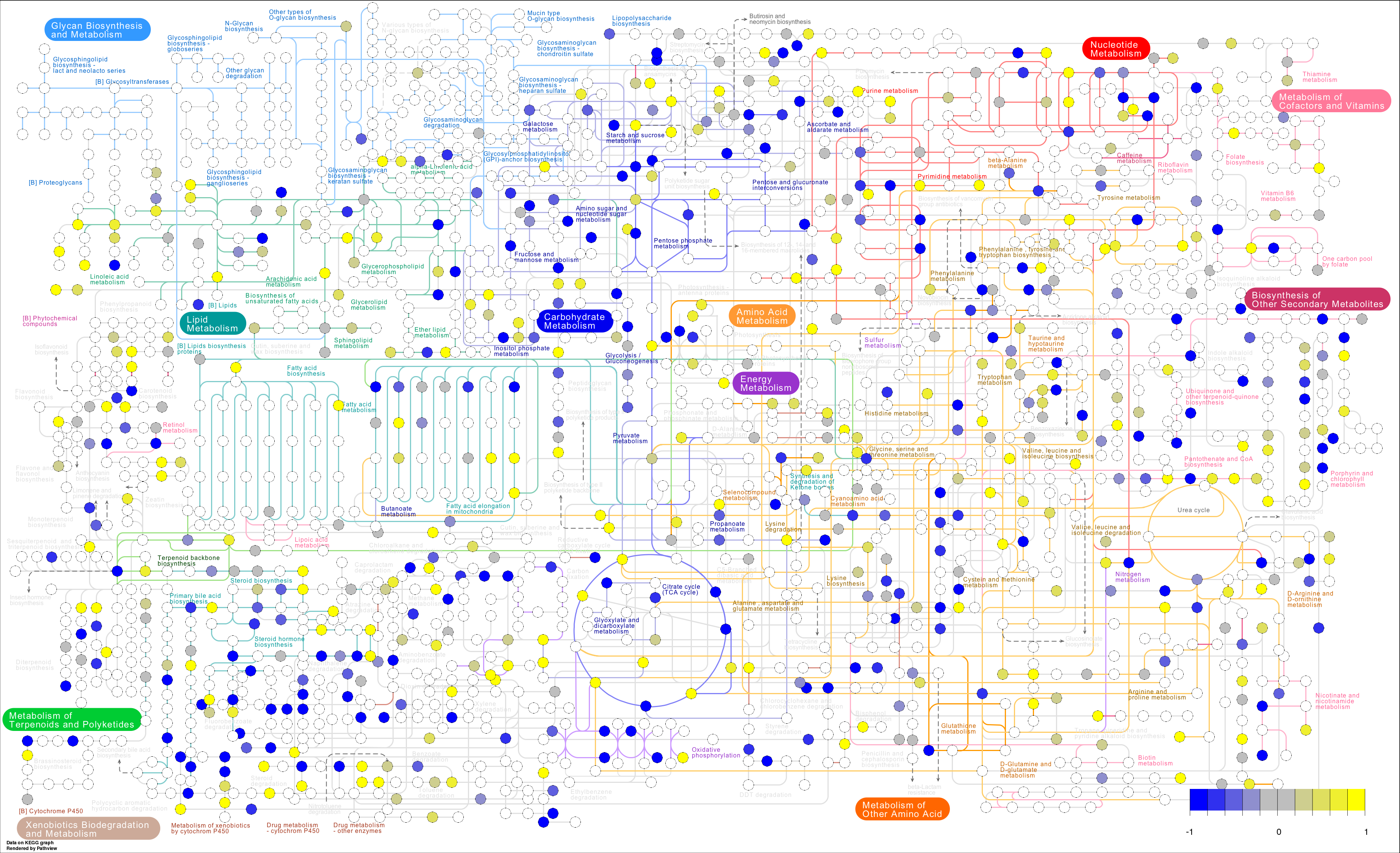
However, I am getting empty circles (all white) and my pathview output file has "NA" for all mol.data and "#FFFFFF" for mol.col
I assume that I am just inputting my data fundamentally incorrectly but I have tried searching the vignette, posts, and doing everything I can think of and I cant figure out why my circles wont get colored in.
This is my code:
temp<-pathview(gene.data = df_pathview[,2],gene.idtype="KEGG", pathway.id = "01100", species = "hsa")
This is how my gene.data df looks:
head(df_pathview)
gene average.mean.diff
3706 ITPKA -1.4976111
2686 GGT7 -8.0186713
216 ALDH1A1 -10.0488333
As you can see my rownames are the entrezIDs and column 2 is my effect sizes (numerics ranging from - to +). Are my entrez ID's not the write IDs to input? I got them with this code:
genekegglinks=getGeneKEGGLinks(species.KEGG = "hsa",convert=FALSE)
Any help would be greatly appreciated as I'm dying to get this metabolic map to work!
Here is my session info:
sessionInfo() R version 4.0.2 (2020-06-22) Platform: x86_64-pc-linux-gnu (64-bit) Running under: Ubuntu 18.04.5 LTS
Matrix products: default BLAS: /usr/lib/x8664-linux-gnu/blas/libblas.so.3.7.1 LAPACK: /usr/lib/x8664-linux-gnu/lapack/liblapack.so.3.7.1
locale:
[1] LCCTYPE=enAU.UTF-8 LCNUMERIC=C
[3] LCTIME=enAU.UTF-8 LCCOLLATE=enAU.UTF-8
[5] LCMONETARY=enAU.UTF-8 LCMESSAGES=enAU.UTF-8
[7] LCPAPER=enAU.UTF-8 LCNAME=C
[9] LCADDRESS=C LCTELEPHONE=C
[11] LCMEASUREMENT=enAU.UTF-8 LC_IDENTIFICATION=C
attached base packages: [1] parallel stats4 stats graphics grDevices utils datasets [8] methods base
other attached packages:
[1] annotate1.66.0 XML3.99-0.5 org.Hs.eg.db3.11.4
[4] AnnotationDbi1.50.3 IRanges2.22.2 S4Vectors0.26.1
[7] Biobase2.48.0 BiocGenerics0.34.0 limma3.44.3
[10] pathview1.28.1 readxl1.3.1 dplyr1.0.2
[13] biomaRt_2.45.5
loaded via a namespace (and not attached):
[1] KEGGgraph1.48.0 Rcpp1.0.5 prettyunits1.1.1
[4] png0.1-7 Biostrings2.56.0 assertthat0.2.1
[7] digest0.6.25 BiocFileCache1.12.1 R62.4.1
[10] cellranger1.1.0 RSQLite2.2.1 httr1.4.2
[13] ggplot23.3.2 pillar1.4.6 zlibbioc1.34.0
[16] rlang0.4.8 progress1.2.2 curl4.3
[19] rstudioapi0.11 Rgraphviz2.32.0 blob1.2.1
[22] stringr1.4.0 munsell0.5.0 RCurl1.98-1.2
[25] bit4.0.4 compiler4.0.2 pkgconfig2.0.3
[28] askpass1.1 openssl1.4.3 tidyselect1.1.0
[31] KEGGREST1.28.0 gridExtra2.3 tibble3.0.3
[34] viridisLite0.3.0 fansi0.4.1 crayon1.3.4
[37] dbplyr1.4.4 bitops1.0-6 rappdirs0.3.1
[40] grid4.0.2 gtable0.3.0 xtable1.8-4
[43] lifecycle0.2.0 DBI1.1.0 magrittr1.5
[46] scales1.1.1 graph1.66.0 cli2.0.2
[49] stringi1.5.3 XVector0.28.0 viridis0.5.1
[52] xml21.3.2 ellipsis0.3.1 generics0.0.2
[55] vctrs0.3.4 tools4.0.2 bit644.0.5
[58] glue1.4.2 purrr0.3.4 hms0.5.3
[61] colorspace1.4-1 memoise1.1.0

