Entering edit mode
ahmad.moousavi
•
0
@ahmadmoousavi-14701
Last seen 6.5 years ago
Hi
I have several questions and I appreciate for answer :
1- I wanted to know how can I add cluster analysis like below image based on read count.
2- Should I run the code only for the genes with minimum number of reads (e.g rowSum(gene) > 10) or I should run it only for DEGs ?
Tnx.
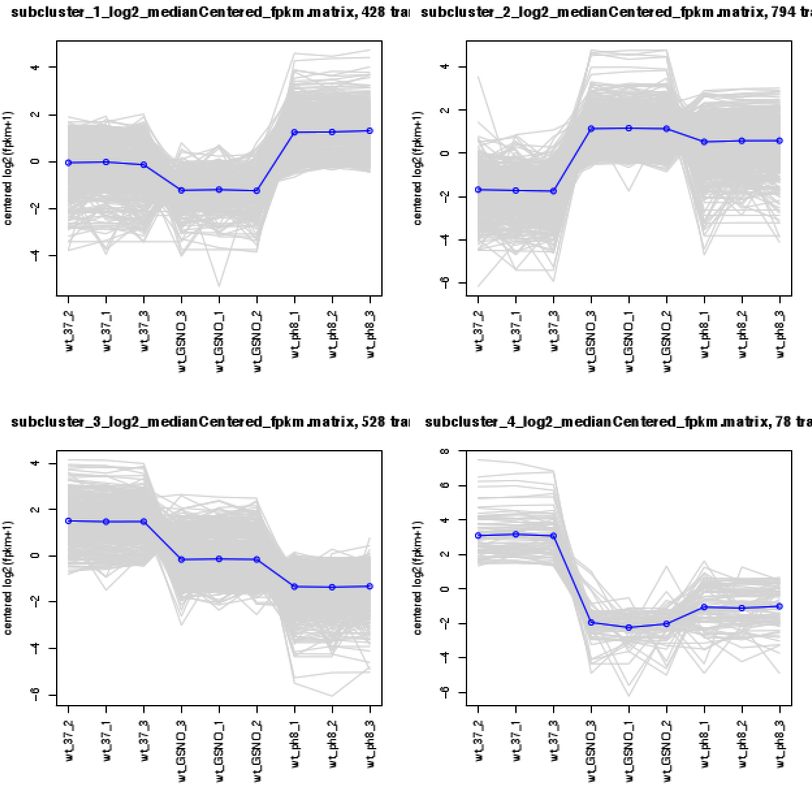


Thanks Michael, at last one answered.
Would you please share your R code with me?
I used to remove low counts by rowSum > 10 is it working too?
I have no code for this.
It’s not built in to DESeq2 but you can use hclust and other base R functions.
thanks, but I need the code !!, I will found the way.
Thanks anyway.