Table 2 of the annual report includes number of packages for each release; there have always been two (spring / fall) releases, but dates before are not readily available (?)
http://bioconductor.org/packages/bioc/1.5/src/contrib/PACKAGES and forward are 'dcf' files produced by the last build of each release -- read.dcf(url("http...")).
http://bioconductor.org/packages/1.8/bioc/VIEWS and forward contain more information, including biocViews terms (biocViews were introduced in June, 2005, I think).
More information about release dates might be obtained by scraping the svn log at https://hedgehog.fhcrc.org/bioconductor/trunk/madman/Rpacks and from the mailing list archives https://hypatia.math.ethz.ch/pipermail/bioconductor and bioc-devel ; the bioconductor mailing list was transferred to the support site, so some creative googling e.g, site:support.bioconductor.org "release date" may lead to additional information, e.g., Release 1.4 information ; I also had success with the support site search engine "release 1.1", etc.
I'm not really sure what you mean by 'views' (maybe biocViews, available from the VIEWS file?); there are download statistics at bioc_pkg_stats.tab available from http://bioconductor.org/packages/stats/ since Jan, 2009.
If you do uncover links to release-like announcements, dates, and other information that could be added to the release-announcements page feel free to post a pull request to https://github.com/Bioconductor/support.bioconductor.org

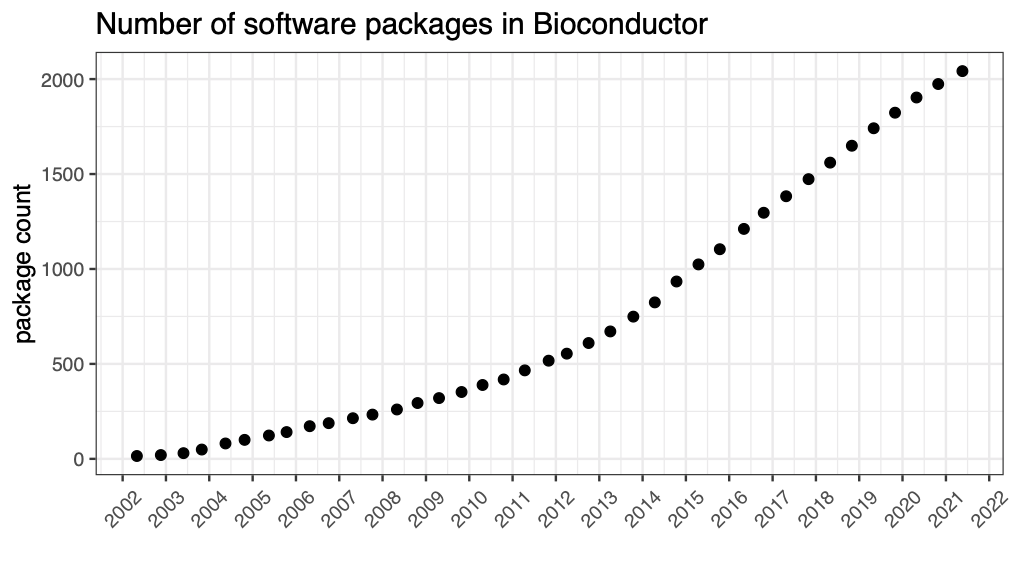


Also not a programmatic solution, but at each release we post the release date and number of packages on the Bioconductor Wikipedia page
Dear Martin and Peter,
Thanks a lot for your helpful answers.
Best, Robert